The human gut is a complex system which consists of not only the host, but myriads of tiny symbionts, commensals, and pathogens, as well as the compounds in the ingested food. The interactions between those elements are not trivial to disentangle. Despite much effort, the full potential of understanding the microbiome has not yet been explored.
MNEMONIC is a method to characterize “shifts” in the microbiome and apply it to study type 2 diabetes (T2D), using thousands of samples available from the American Gut Project (AGP) repository. We aim to compare changes observed between experiment and control conditions, so that similar “shifts” can be identified. For example the changes observed between a healthy person and a diabetic patient might be similar to the changes in different experiments where subjects went from condition A to condition B. Alternatively, shifts might be similar to those observed under different dietary habits.
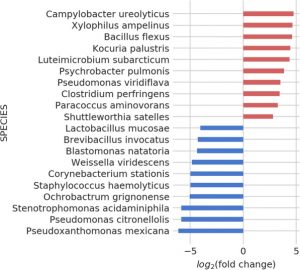
We perform a differential abundance analysis between samples from diabetic and non-diabetic people. A distance matrix between vectors of these changes serves as a basis for this part of the analysis. We also cross-referenced taxa with literature terms, as well as a microbial annotation database ProTraits, which allows wider conclusions to be drawn by extending the pool of available terms.
Figure: Changes in microbial species in autism spectrum disorder patients